Medical Management
Identifying Ventricular Tachycardia Events in Unstructured Medical Notes using Natural Language Processing
Ruibin Feng, PhD – Basic Life Research Scientist, Cardiovascular Medicine, Stanford University; Xin Ai, MS – Data Analysis II, Cardiovascular Medicine, Stanford University; Jatin Goyal, BS – Research Associate, Cardiovascular Medicine, Stanford University; Zahra Azizi, MD – Postdoctoral Scholar, Cardiovascular Medicine, Stanford University; Paul Clopton, MS – Statistician, Cardiovascular Medicine, Stanford University; Brototo Deb, MD – Internal Medicine Resident, Georgetown University; Prasanth Ganesan, PhD – Research Scientist, Cardiovascular Medicine, Stanford University; Hui Ju Chang, PhD, PT – Clinical Research Coordinator, Cardiovascular Medicine, Stanford University; Samuel Ruiperez-Campillo, MSc, MEng – Research Associate, Cardiovascular Medicine, Stanford University; Maxime Pedron, MS – Research Associate, Cardiovascular Medicine, Stanford University; Tina Baykaner, MD, MPH – Assistant Professor of Medicine, Cardiovascular Medicine, Stanford University; Albert Rogers, MD, MBA – Early-career Physician, Cardiovascular Medicine, Stanford University; Sanjiv Narayan, MD, PhD – Professor of Medicine, Cardiovascular Medicine, Stanford University
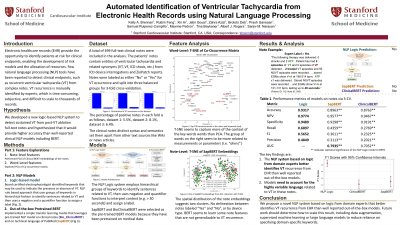
Purpose: Tools that can automatically interpret Electronic Healthcare Record (EHR) systems to identify ventricular tachycardia (VT) recurrence could be useful to patients, to third party agents developing diagnostic apps, to researchers, and for quality assurance tasks by administrators. However, expert chart review remains the gold standard to interpret complex, unstructured EHR notes, which is time consuming, and does not facilitate automated diagnostics. We hypothesized that a logic-based natural language processing (NLP) system to detect recurrent VT in clinical notes would outperform available state-of-the-art models.
Material and Methods: We studied N=499 full-text clinical notes (474.6 ± 164.3 words) in N = 125 subjects (32.0% female, LVEF 48.9 ± 13.9%, 61 ± 14.0 years) from our VT ablation registry. Experts reviewed each note in context and labeled them as “Yes” or “No” for the nuanced endpoint of recurrent VT after ablation, filtering out mentions of VT prior to ablation. We developed an NLP system based on hierarchical groups of keywords which we compared to reported models (clinicalBERT and SapBERT).
Results: The bespoke NLP system improved upon models pre-trained for other domains, achieving 94.5% accuracy (56.8% precision, 74.9% recall) versus SapBERT ( 91.0% accuracy; 29.1% precision, 31.0% recall; p< 0.05) or clinicalBERT (87.6% accuracy, 16.6% precision, 27.7% recall; p< 0.05). Approaches to improve BERT, such as fine-tuning, did not improve their performance relative to the domain-specific NLP (p>0.05) .
Conclusions: Domain-specific NLP systems identified VT recurrence from electronic health records significantly better than state-of-the-art pre-trained BERT models. This approach may enable novel diagnostic apps for various domains, could enable smaller institutions to provide quality assurance, could be used by researchers to identify at-risk patients, and could potentially study and reduce biases from manual labeling.
Material and Methods: We studied N=499 full-text clinical notes (474.6 ± 164.3 words) in N = 125 subjects (32.0% female, LVEF 48.9 ± 13.9%, 61 ± 14.0 years) from our VT ablation registry. Experts reviewed each note in context and labeled them as “Yes” or “No” for the nuanced endpoint of recurrent VT after ablation, filtering out mentions of VT prior to ablation. We developed an NLP system based on hierarchical groups of keywords which we compared to reported models (clinicalBERT and SapBERT).
Results: The bespoke NLP system improved upon models pre-trained for other domains, achieving 94.5% accuracy (56.8% precision, 74.9% recall) versus SapBERT ( 91.0% accuracy; 29.1% precision, 31.0% recall; p< 0.05) or clinicalBERT (87.6% accuracy, 16.6% precision, 27.7% recall; p< 0.05). Approaches to improve BERT, such as fine-tuning, did not improve their performance relative to the domain-specific NLP (p>0.05) .
Conclusions: Domain-specific NLP systems identified VT recurrence from electronic health records significantly better than state-of-the-art pre-trained BERT models. This approach may enable novel diagnostic apps for various domains, could enable smaller institutions to provide quality assurance, could be used by researchers to identify at-risk patients, and could potentially study and reduce biases from manual labeling.
